Table Of Contents:
The False Dichotomy: Structured vs. "Unstructured" Data
The Power of Shape-Shifting Arrays
Building Databases, Not Formats
The Technical Foundation
Why This Matters for Scientific Discovery
A New Approach to Data Management
In the world of life sciences research, we face a fundamental data challenge: how do we efficiently store, access, and analyze the diverse data modalities driving modern discovery? From genomic variants and single-cell transcriptomics to proteomics and biomedical imaging, each data type seems to demand its own specialized format and tooling. But what if this fragmentation isn't necessary? What if we've been thinking about data structure all wrong?
The False Dichotomy: Structured vs. "Unstructured" Data
Consider a simple thought experiment: imagine searching for a specific number in a large, randomly ordered list. Your computer would need to scan the entire list, checking each value one by one. Now, imagine the same list, but sorted. Suddenly, your computer can use binary search, dramatically reducing the number of operations needed to find your value.
This example illustrates a critical insight: computational efficiency depends fundamentally on how data is structured. Yet the market has created an artificial dichotomy, categorizing data as either "structured" (tabular) or "unstructured" (everything else).
This categorization has led to a problematic approach in life sciences. Tabular data gets efficient database treatment, while complex data like genomic variants, single-cell matrices, and imaging is relegated to bespoke file formats lacking database features. These formats are rarely cloud-optimized, resistant to change, and create data silos that hinder discovery.
But here's the truth: no data is truly unstructured. An image isn't a random collection of pixels; it's a precise 2-D matrix with well-defined relationships. Genomic variants have clear positions and relationships. The challenge isn't that these data lack structure; it's that we've lacked a universal data structure flexible enough to capture their diverse organizations efficiently.
The Power of Shape-Shifting Arrays
At TileDB, we've architected our entire data engine around a powerful mathematical concept: the multi-dimensional array. Arrays are more than just rigid grids of numbers, they're shape-shifting data structures that can adapt to represent virtually any data type, no matter how complex.
The power of this approach stems from two fundamental concepts that are at the heart of efficient data management:
- 1
Optimized data layout: Arrays allow us to organize data values in specific patterns on storage media, making search operations dramatically more efficient.
- 2
Sophisticated indexing: Arrays support powerful indexing structures built on top of the data values, expediting search operations without requiring full scans.
These concepts, which have been central to database performance for decades, can now be applied to all data types through the flexibility of arrays, which come in two fundamental varieties:
Dense arrays: Every cell contains a value, perfect for data like images where each pixel has information
Sparse arrays: Only non-empty cells store values, ideal for naturally sparse data like single-cell matrices
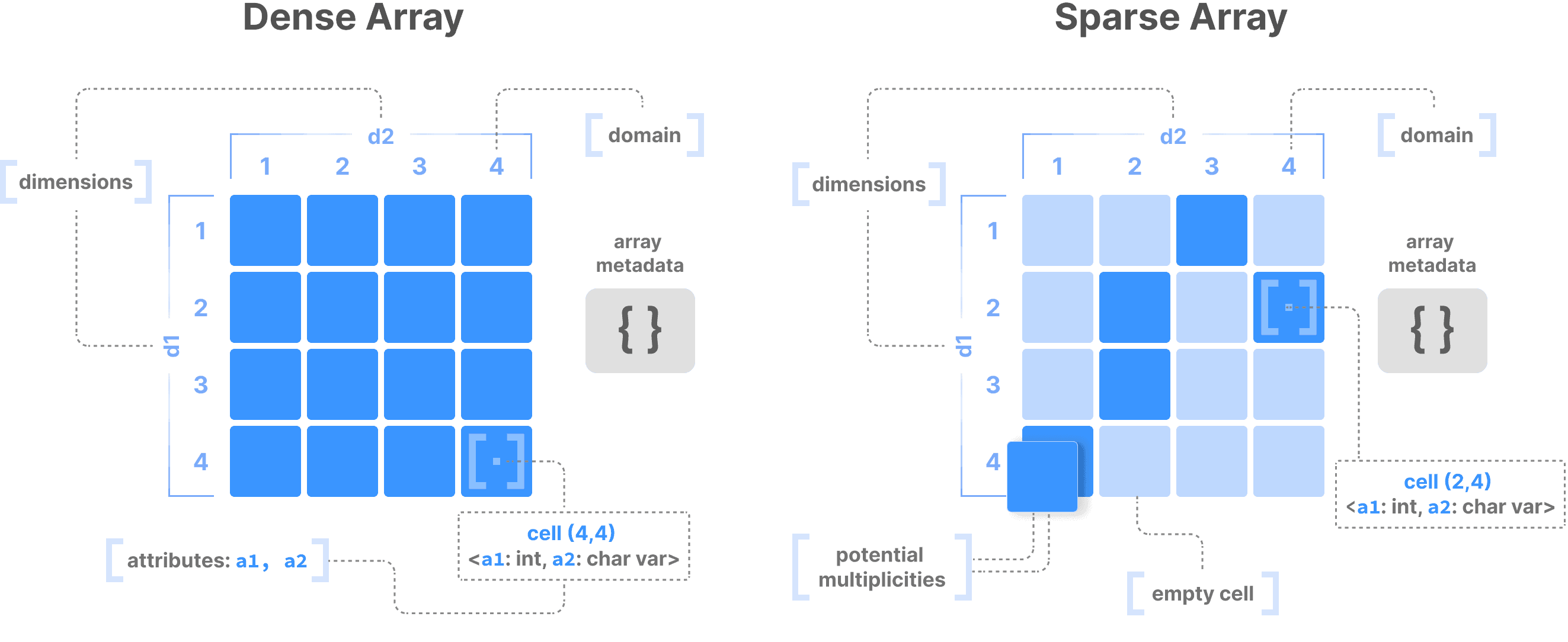
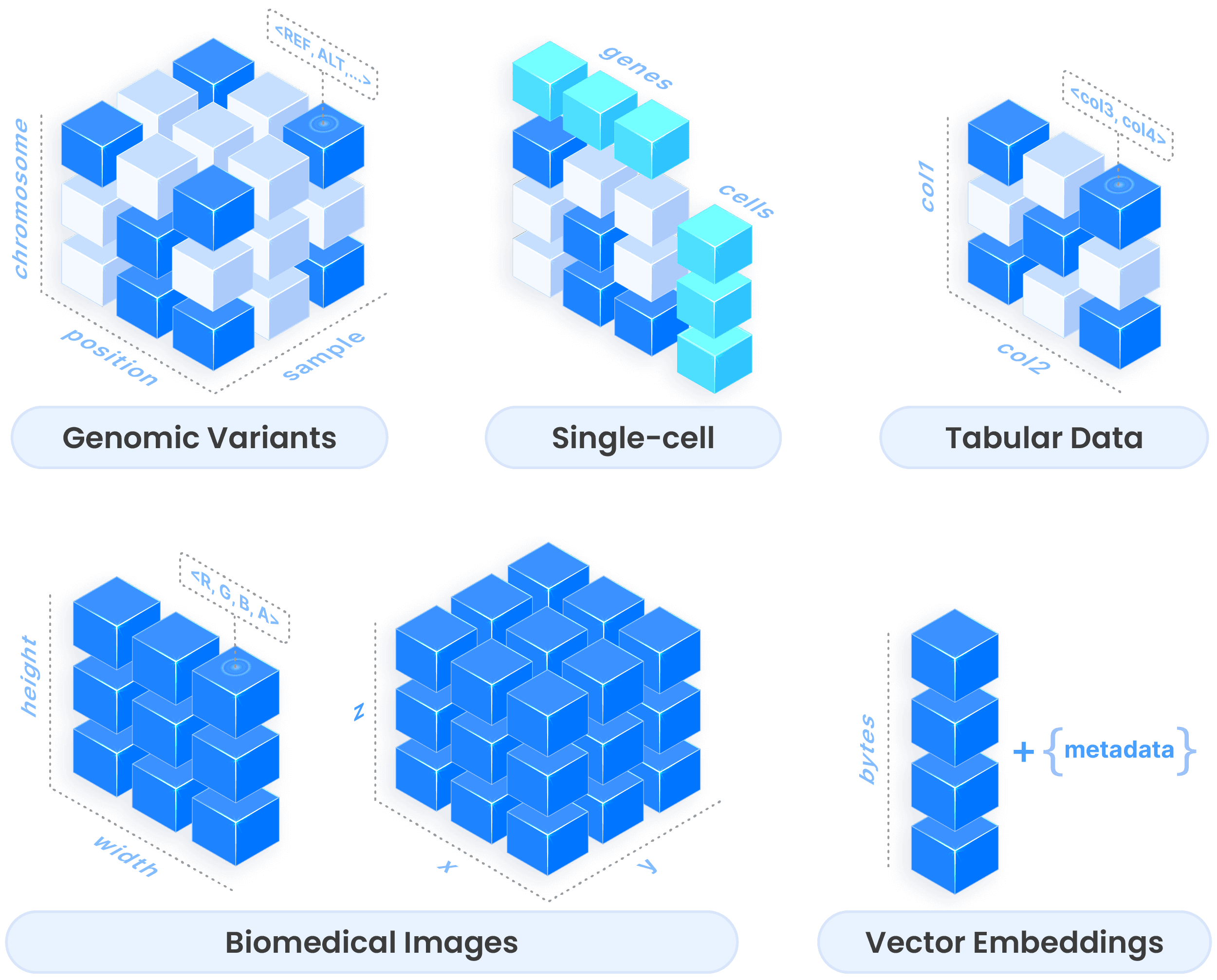
This simple distinction enables tremendous flexibility. Here's how arrays shape-shift to accommodate diverse data types:
Genomic variants are modeled as sparse 3-D arrays, efficiently representing variant positions and alt/ref alleles
Single-cell data uses the SOMA format, a collection of arrays and groups that perfectly capture cell-by-gene matrices
Biomedical images become 2-D or 3-D dense arrays, with attributes for color channels
Tabular data becomes a specialized case of arrays, where dimensions act as indices and attributes store column values
Vector embeddings for AI applications are simply 1-D dense arrays
Building Databases, Not Formats
The shape-shifting array model represents a fundamental shift from creating bespoke file formats to building true database systems for complex data. While traditional file formats focus on how bytes are laid out, TileDB focuses on standardizing APIs and access patterns — similar to how SQL standardized tabular data access regardless of the underlying implementation.
Cloud-optimized performance: Arrays are tiled and can be accessed without downloading entire datasets
Transactional guarantees: Multi-reader/multi-writer support with versioning
Performant queries: Efficient slicing, filtering, and aggregation across all modalities
Secure collaboration: Fine-grained access controls and audit logging
Standardized APIs: Consistent access patterns across all data types
The Technical Foundation
The technical implementation of arrays in TileDB offers powerful solutions for life sciences challenges. For sparse data like single-cell matrices, R-tree indexing enables efficient region queries while avoiding the storage of empty values. For dense data like imaging, implicit indexing allows direct calculation of cell positions without separate index structures.
Most importantly, by structuring all data, regardless of modality, using the same fundamental array model, TileDB enables seamless integration across previously siloed data types. A researcher can query genomic variants alongside transcriptomic data and imaging in a unified computational framework, opening new possibilities for integrative analysis.
Why This Matters for Scientific Discovery
The technical architecture underlying scientific data systems isn't just an engineering concern; it directly determines which scientific questions can be feasibly asked and answered at scale. With multimodal multiomics now driving drug discovery, having a unified data foundation becomes essential.
By removing the arbitrary boundary between "structured" and "unstructured" data, shape-shifting arrays transform what were once insurmountable technical challenges into routine operations. Scientists can focus on biology rather than data engineering, enabling:
- 1
Elimination of silos: Seamless integration across data modalities
- 2
Performance at scale: Efficient handling of biobank and atlas-scale datasets
- 3
Enhanced collaboration: Secure data sharing with detailed governance
- 4
Integrated analysis: Cross-modal queries that were previously impractical
A New Approach to Data Management
Modern life sciences research demands a new approach to data management — one that recognizes the inherent structure in all data types and provides a unified computational framework for analysis at scale.
By leveraging the shape-shifting capabilities of multi-dimensional arrays, TileDB offers a solution that transcends the limitations of traditional file formats and database systems. It's not just about storing data more efficiently, it's about enabling new classes of scientific questions that drive discovery.
The next time someone mentions "unstructured" data, remember: it's not that the data lacks structure, we just needed a better way to capture and leverage that structure. With shape-shifting arrays, we can finally do justice to the rich complexity of multimodal multiomics data, accelerating the path from data to insight to impact.
Meet the authors

