Table Of Contents:
Managing Diverse Data Types in One Environment
From Reference Files to Queryable Databases
Nextflow Workflows: From Sequence to Database
Debugging and Monitoring Workflows
From Workflow Results to Interactive Analysis
The Analytic Stack: From Daily Driver to Trusted Research Environment
Want to Transform Your Analysis Workflow?
Watch the Video: TileDB Carrara Demo | From Raw Data to Reproducible Results
In our latest video, we demonstrate how TileDB Carrara addresses these challenges by providing a unified platform where every step of the analysis workflow happens in one integrated environment. From organizing reference files to running Nextflow pipelines to interactive data exploration, Carrara streamlines the analyst's daily workflow while maintaining the rigor and compliance requirements of modern bioinformatics. In this article, I’ll walk through the workflow demonstrated in the video.
Managing Diverse Data Types in One Environment
Anyone working in the life sciences world faces an overwhelming diversity of multimodal data. Multiomics analysis alone requires managing raw sequence data, alignment files, reference genomes, gene annotations, TileDB-SOMA arrays for expression data, quality control reports, and analysis notebooks. In traditional workflows, these assets live in disconnected systems, each requiring different tools and access patterns.
Carrara treats all of these assets as first-class citizens within a unified catalog. Genomic sequences, TileDB-VCF variant stores, TileDB-SOMA single-cell databases, imaging data, reports, and notebooks all live together in one searchable environment. The ability to add rich metadata and descriptions to any asset powers advanced search capabilities and makes data discovery intuitive and less frustrating.
Teamspaces: Secure Collaboration Made Simple
Teamspaces provide project-level environments where teams manage permissions and security for data access. When files are mapped into a teamspace, Carrara handles all the cloud security wiring, including identity and access management, audit logging, and compliance tracking, without moving or copying your data.
This logical mapping approach is crucial for efficiency. Files can exist in multiple teamspaces without bloating storage with redundant copies. Adding collaborators is as simple as inviting them to a teamspace and setting appropriate permissions: read-only viewer, editor with write access, or owner with administrative rights. All actions are audit-logged for compliance requirements. You can learn more about Teamspaces in our article: From file chaos to unified data discovery.
From Reference Files to Queryable Databases
A typical genomics analysis begins with organizing reference files. In this workflow, we can register genome reference sequences and gene annotation files from any cloud provider and object store.
In Carrara, files have familiar folder hierarchies and file operations, but with added capabilities: searchable metadata, programmatic API access, secure sharing, and governance. Carrara creates logical mappings rather than copying data, so storage costs can be kept to a minimum, even across large projects.
We can enrich these reference files with custom metadata tags, adding descriptions, along with tags for genome build and annotation version. This metadata makes files discoverable later when setting up workflows or sharing resources with collaborators.
Nextflow Workflows: From Sequence to Database
Secondary analysis, the long-running file-based transformations typical in bioinformatics, has been revolutionized by pipeline frameworks like Nextflow. Carrara now provides native support for Nextflow workflows, making it appealing for both power users who prefer command-line interfaces and end users who benefit from visual workflow management.
TileDB has developed Nextflow modules specifically for TileDB-VCF and TileDB-SOMA that allow users to go directly from raw sequence data to queryable databases. These modules integrate seamlessly with popular community workflows like those from nf-core, a curated collection of bioinformatics pipelines.
Interactive Workflow Configuration
Workflows that follow the nf-core specification can be rendered as interactive forms in Carrara's dashboard interface. Analysts can import workflows from nf-core directly, from GitHub repositories, or even upload custom workflow definitions from local systems.
In this example, we import a TileDB-specific fork of the popular nf-core scrnaseq workflow. This workflow produces TileDB-SOMA assets directly from raw sequence data in a single run, eliminating manual conversion steps and ensuring data format consistency.
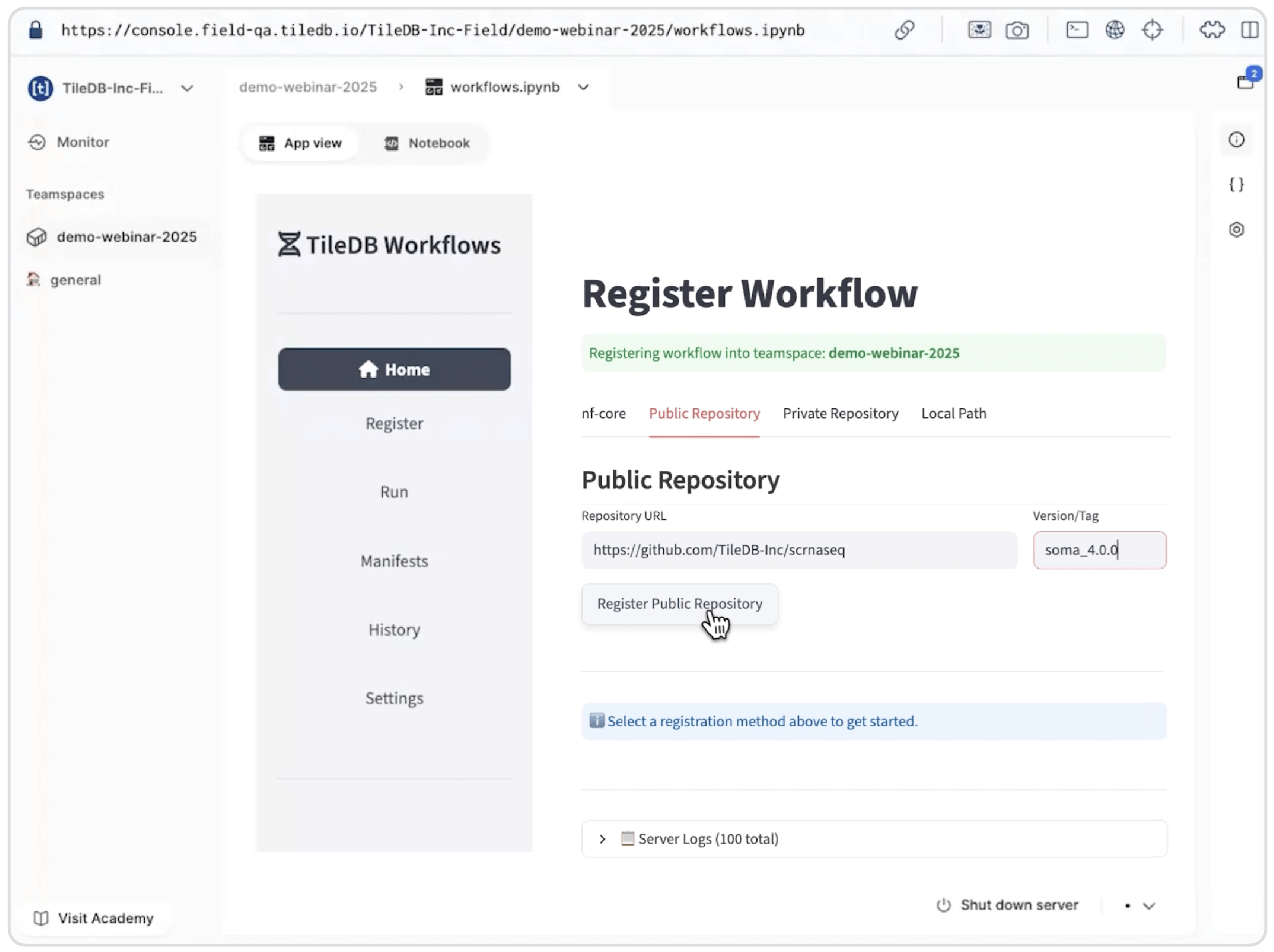
Importing a TileDB-specific fork of the nf-core scrnaseq workflow from GitHub, specifying version soma_4.0.0 to generate TileDB-SOMA assets directly from raw sequencing data.
Once imported and registered, the workflow presents all configuration options through an intuitive form interface. The README documentation, including usage instructions, appears directly in the interface. Form elements map to workflow parameters, making it easy to specify inputs like sample sheets, sequencing protocols (10X Chromium V3), alignment tools (STAR), and critically, reference files already registered in the TileDB filesystem.
The workflow configuration references files using TileDB uniform resource identifiers (URIs), like tiledb://TileDB-Inc-Field/demo-webinar-01/Homo_sapiens.GRCh38.dna_sm.primary_assembly.fa, creating a direct connection between the catalog and computational workflows. This tight integration ensures reproducibility and eliminates the common problem of mismatched references or lost file paths.
Workflow Manifests: Reusability and Provenance
This creates a provenance chain: workflows link to manifests, manifests link to runs, and runs link to results. When reviewers or collaborators need to understand how data was generated, they can trace the entire analysis linearly through these connections.
When the analyst kicks off a run, Carrara uses the same infrastructure that powers TileDB's task graphs for tertiary analysis. This means workflows benefit from the same debugging tools, resource management, and monitoring capabilities available for other computational tasks.
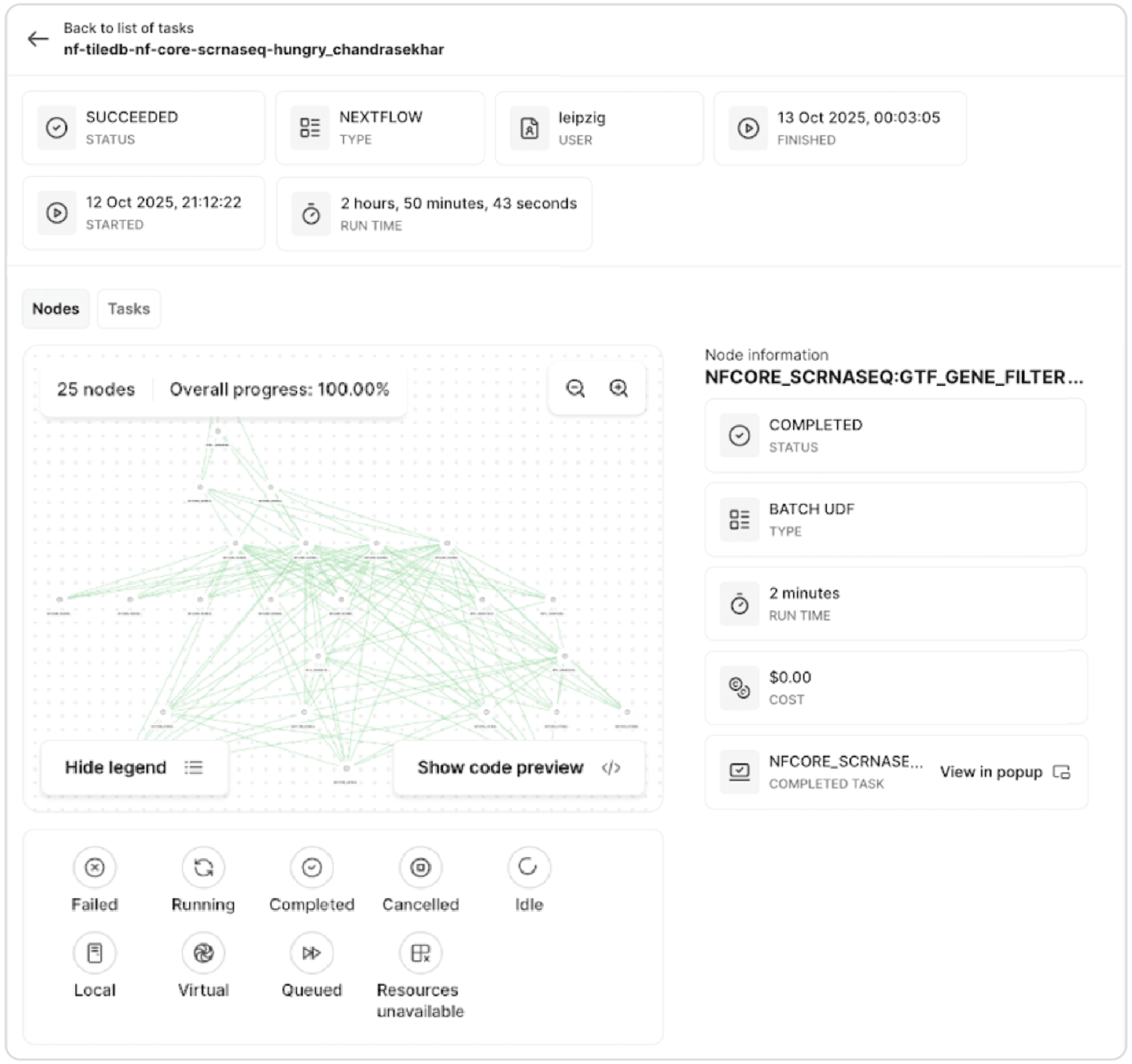
Task graph visualization showing a running Nextflow workflow with three nodes and individual task details, including status, type, runtime, and cost metrics.
Debugging and Monitoring Workflows
Carrara is built not just for running workflows, but for debugging them. The task graph visualization shows individual workflow tasks, their status (i.e., succeeded, running, failed), and their logging output. Analysts can inspect why specific tasks failed and adjust resource specifications without rerunning the entire pipeline.
From Workflow Results to Interactive Analysis
When a workflow completes, Carrara provides immediate access to results through built-in preview capabilities. HTML and markdown reports render securely in the browser, even if they contain JavaScript elements. This includes Nextflow's execution reports, which show computational efficiency metrics like CPU usage, memory consumption, and task duration, providing critical information for optimizing future runs.
MultiQC reports, which aggregate quality control metrics across samples, also render directly in the interface. Analysts can evaluate alignment statistics, sequencing quality, and other pipeline outputs without downloading files or switching to external tools.
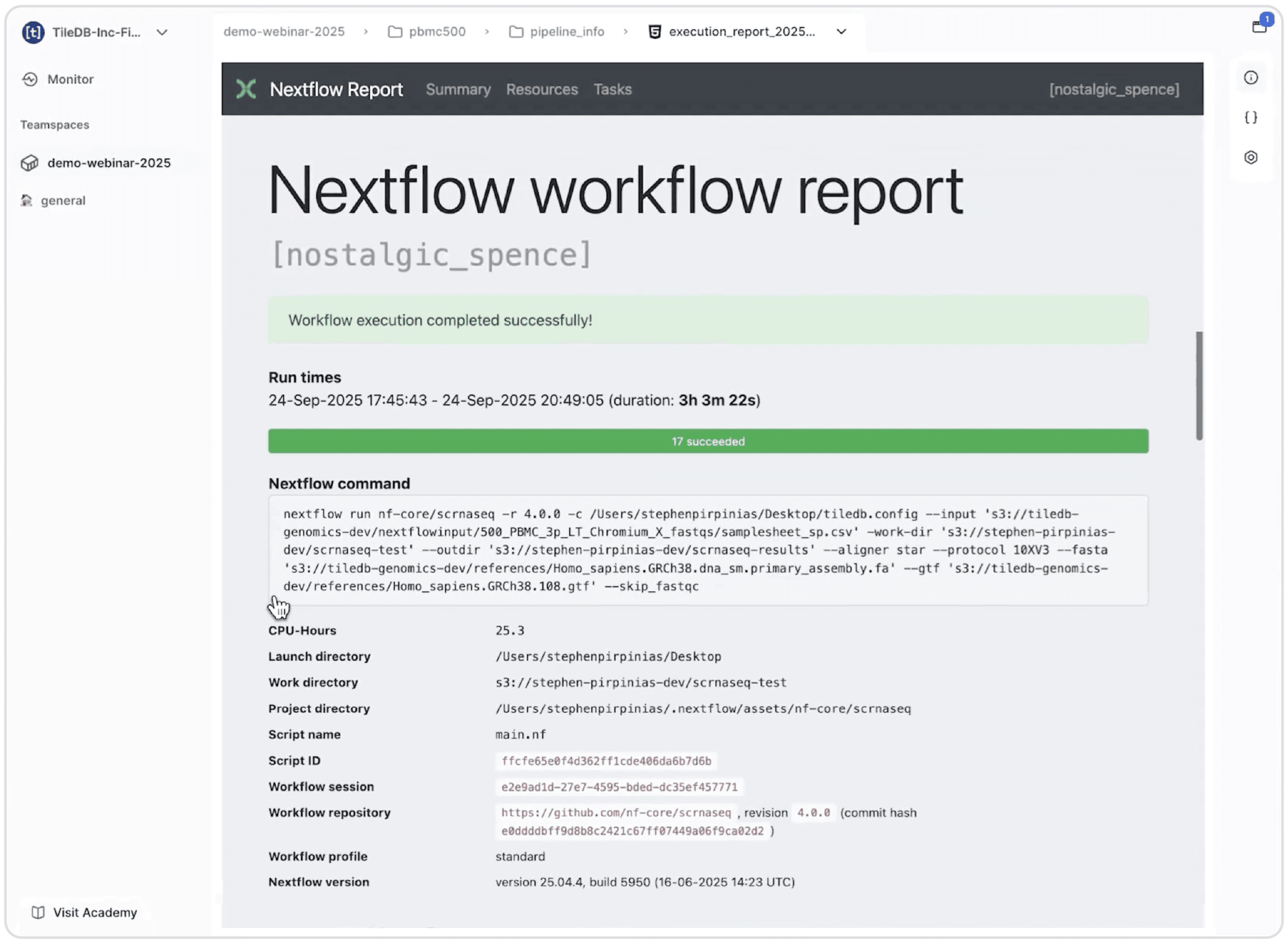
Nextflow execution report is rendered directly in Carrara's browser interface, showing workflow completion status, runtime duration, successful tasks, and detailed execution parameters including CPU usage and file paths.
Tertiary Analysis with Notebooks
The workflow produces TileDB-SOMA output: a specialized array format optimized for single-cell expression data. We can immediately query this data in Carrara's revamped notebook environment, which provides both traditional notebook interfaces and streamlined dashboard views.
The notebook environment includes a tightly integrated terminal that provides direct access to teamspace files. In this example, we download cell type marker files from the Human Protein Atlas directly from the terminal. Files created or downloaded in the terminal immediately appear in the catalog, maintaining a unified view of all analysis assets.
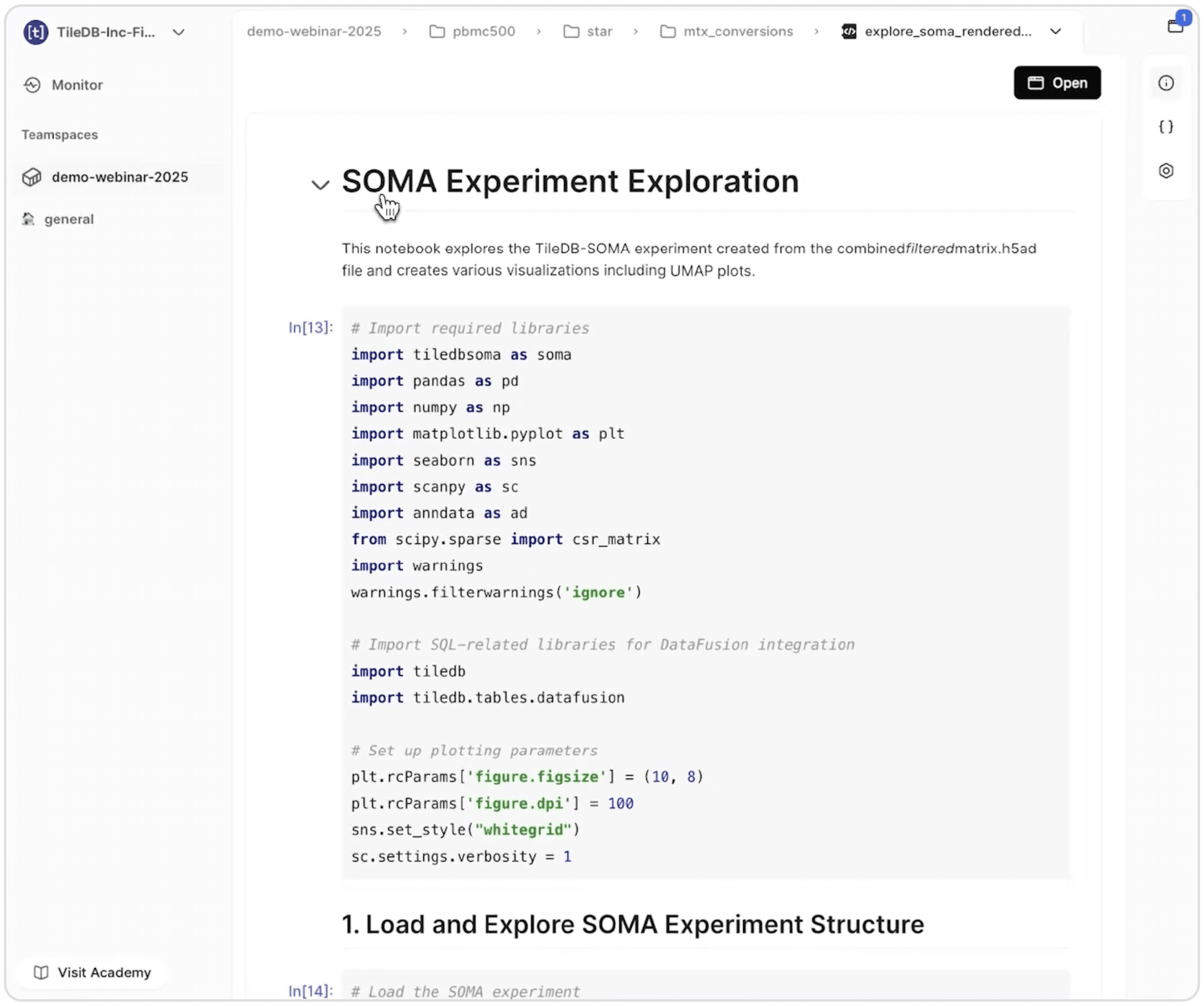
Carrara's notebook interface shows a SOMA Experiment Exploration notebook with Python code for importing analysis libraries including tiledbsoma, pandas, and visualization tools like matplotlib and seaborn for single-cell data analysis.
High-Performance Table Operations
While TileDB is renowned for handling multi-dimensional arrays and complex data structures, Carrara also excels at traditional table operations. The platform now uses Apache DataFusion with Arrow in-memory representation, providing high-performance table queries with a Rust-based backend.
Analysts can write SQL directly against TileDB-SOMA tables, running aggregations and transformations on observation (cell) and variable (gene) matrices. The notebook demonstrates summary statistics computed using both SQL and dataframe syntax, showing the flexibility of the platform.
This combination of array-native operations for complex genomics data and high-performance table operations for metadata creates a complete analytical environment. Analysts don't need to export data to specialized tools or switch between platforms, because everything happens in one integrated workspace.
The Analytic Stack: From Daily Driver to Trusted Research Environment
These capabilities — the catalog, teamspaces, filesystem, workflows, tables, and notebooks — work together to create what TileDB calls the analytic stack. This stack is built around the backbone of TileDB's multimodal database, which naturally accommodates the diverse data types and complex structures common in life sciences research.
Carrara serves as both a daily driver for analysis and a Trusted Research Environment (TRE) for secure data sharing. The platform enables different roles to work effectively in their domains: developers manage raw files and pipelines, scientists analyze data in notebooks, and technical leaders create dashboards for stakeholder reporting. Governance ensures all access is logged and auditable, meeting compliance requirements without sacrificing usability.
The seamless handoff from secondary to tertiary analysis, from workflow completion to immediate data exploration, eliminates common bottlenecks in research pipelines. Data doesn't need to be moved, reformatted, or re-ingested. Results flow directly into analysis environments with full provenance tracking and reproducibility built in.
Want to Transform Your Analysis Workflow?
Modern single-cell analysis demands a platform that can keep pace with the complexity and scale of the data. TileDB Carrara provides that platform, unifying data organization, computational workflows, and interactive analysis in one governed environment.
Whether you're managing growing biobank projects with daily sample additions or conducting complex multi-modal analyses across genomics and imaging data, Carrara's N+1 strategy (adding new samples without reprocessing existing data) and multimodal architecture provide the foundation for scalable, reproducible science.
Ready to see how Carrara can streamline your single-cell analysis workflows? Talk to us to schedule a personalized demo and discover how to accelerate your journey from raw sequence to biological insight.
Meet the authors

