Table Of Contents:
Deploy Workflows Your Way
Universal Nextflow Support
Collaborative Workflow Management
Optimized Integration with TileDB Data Formats
Real-Time Monitoring and Resource Tracking
From Rigid Pipelines to Flexible Science
Watch the Video: TileDB Carrara Demo | Run workflows your way
Running bioinformatics pipelines shouldn't mean compromising on how your team works. Yet that's exactly what happens with using traditional workflow management systems with bioinformatic portals. You're locked into specific execution environments, forced to recreate complex configurations when sharing workflows, and required to restart from scratch when jobs fail, wasting compute resources and time.
TileDB Carrara transforms workflow management by giving you complete flexibility in how you run bioinformatics pipelines while maintaining the rigor and reproducibility that scientific computing demands. Whether you prefer command-line execution or graphical interfaces, working solo or collaborating across teams, Carrara adapts to your needs.
Deploy Workflows Your Way
We recognize that different team members have different preferences and expertise levels. Data scientists comfortable with terminal commands can run workflows from the command line on their laptop or cloud instance. Scientists who prefer visual interfaces can use the intuitive dashboard GUI. The same powerful workflows run seamlessly across both interfaces.
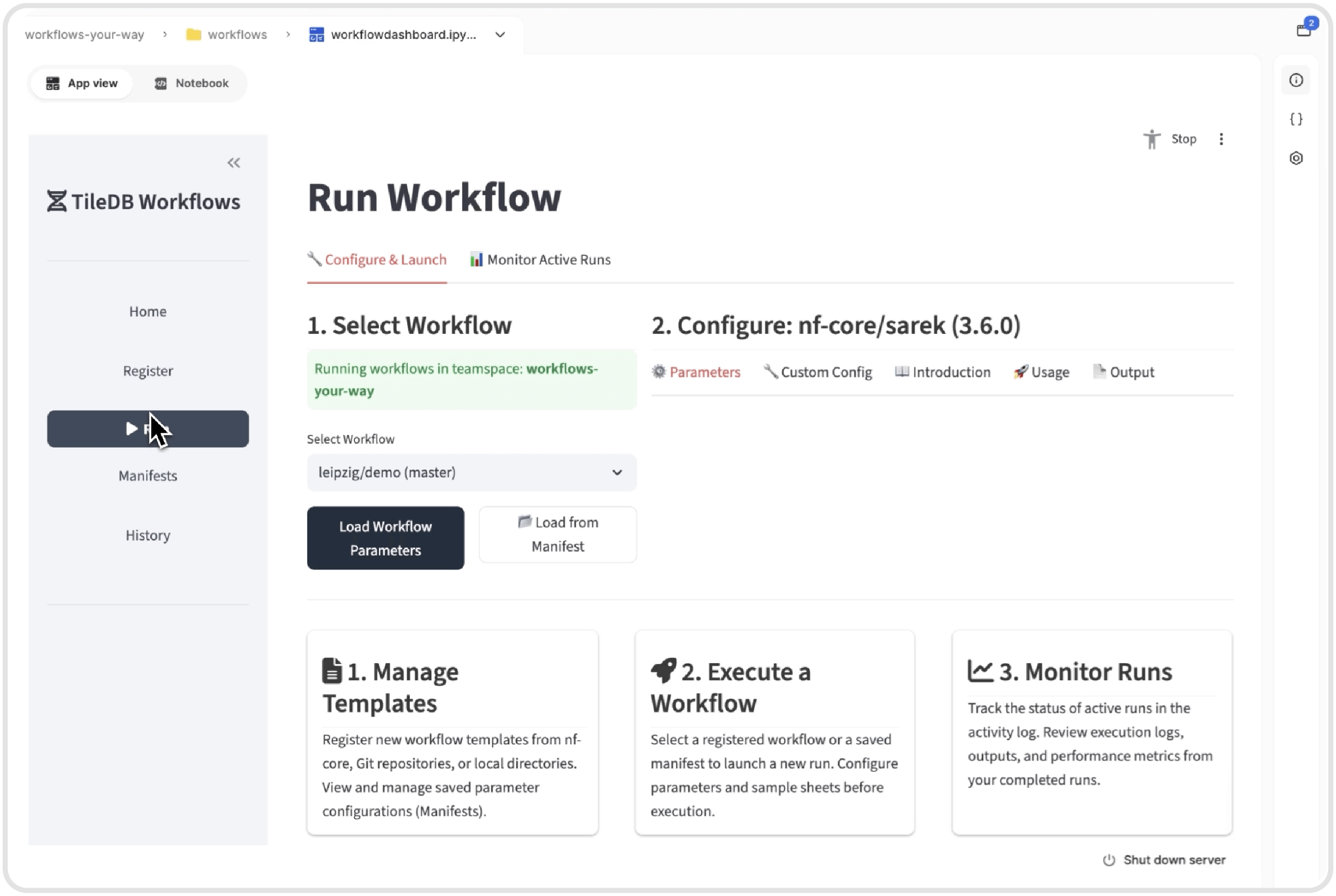
Running a workflow in TileDB Carrara can be done using the dashboard or terminal commands on your laptop or cloud instance.
This flexibility eliminates a common bottleneck in research teams: workflow gatekeeping. Junior scientists no longer need to master complex command-line tools to run validated pipelines, while experienced bioinformaticians maintain full control through their preferred terminal environment.
Universal Nextflow Support
The Carrara command-line interface supports any Nextflow workflow without modification. Treat TileDB as just another runner, and your existing pipelines work immediately. No special adaptations, no vendor lock-in, no rewriting your carefully tested code.
For teams using the dashboard interface, Carrara supports nf-core compliant workflow templates. Pull workflows directly from nf-core, GitHub repositories, or upload from your computer. The visual interface removes command-line complexity while maintaining the full power of validated scientific workflows.

The dashboard supports nf-core compliant workflow templates. You can pull directly from nf-core, GitHub, or upload from your computer.
Collaborative Workflow Management
Optimized Integration with TileDB Data Formats
Carrara includes TileDB-optimized modules for popular nf-core pipelines like Sarek and scrnaseq. These modules take you directly from raw sequences to queryable TileDB datasets: TileDB-VCF for genomic variants and TileDB-SOMA for single-cell data.
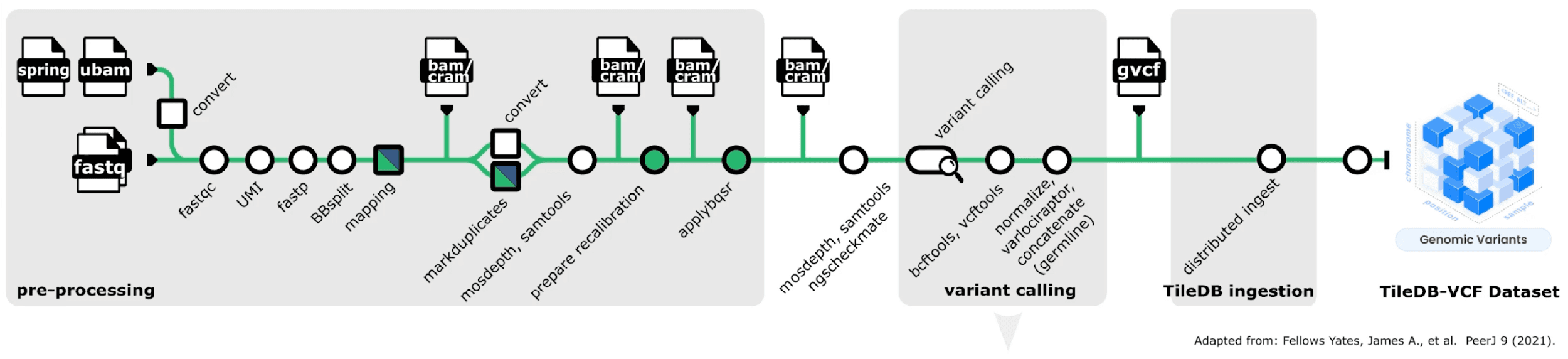
We've created TileDB modules for popular nf-core pipelines like Sarek and scrnaseq. You can go directly from raw sequences to optimized queryable datasets: TileDB-VCF for genomic variants and TileDB-SOMA for single-cell data.
This tight integration eliminates the typical gap between pipeline completion and data analysis. Results land directly in formats optimized for downstream queries, visualization, and analysis within the same Carrara environment.
Real-Time Monitoring and Resource Tracking
Never wonder about job status again. Monitor workflow progress through Nextflow logs in the terminal or watch visual task graphs in the dashboard. Every step is tracked, every resource monitored, and every failure captured for debugging.
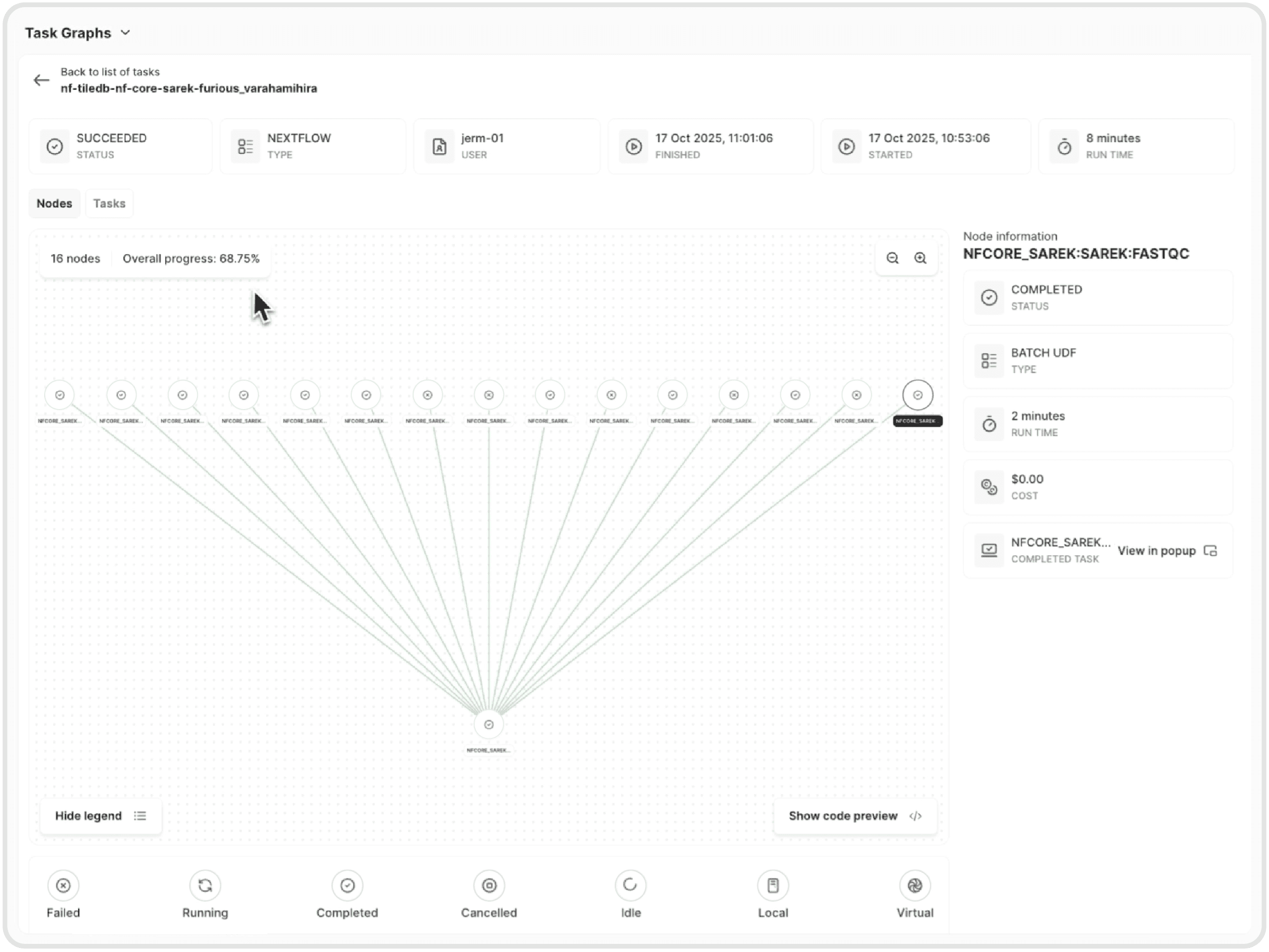
You can monitor progress from the Nextflow logs or watch visual task graphs in the TileDB Carrara dashboard.
When jobs do fail, Carrara's monitoring reveals exactly where and why, enabling targeted troubleshooting rather than blind restarts. Resource monitoring helps teams optimize compute allocation and control costs.
From Rigid Pipelines to Flexible Science
Ready to run workflows your way? Discover the workflow capabilities of TileDB Carrara and start streamlining your bioinformatics pipelines today.
Meet the authors

