Table Of Contents:
What are the FAIR data principles?
Who developed the FAIR data principles?
What does each FAIR principle stand for?
What is the difference between FAIR data and open data?
Why are the FAIR data principles important?
Enables faster time-to-insight
Improves data ROI and reduces infrastructure waste
Supports AI and multi-modal analytics
Ensures reproducibility and traceability
Enables better team collaboration across silos
How do the FAIR principles differ from CARE principles?
What are the common challenges in implementing FAIR data principles?
Fragmented data systems and formats
Lack of standardized metadata or ontologies
High cost and time investment in transforming legacy data
Cultural resistance or lack of FAIR-awareness in teams
Infrastructure built for tabular, not multi-modal, data
How to make data more FAIR?
Findable
Accessible
Interoperable
Reusable
Conclusion: TileDB for your FAIR data solution?
FAIR principles are often discussed alongside open data and CARE principles, but these data rules have key differences. The difference between open and FAIR data is that open data focuses on unrestricted public access and is available to all, while FAIR data is designed for computers to process and defines specific conditions for data to be accessed and used. FAIR principles differ from CARE principles in that FAIR principles aim to facilitate data sharing using technology and CARE principles focus on the key role of data in advancing the innovation and self-determination of Indigenous people.
Common challenges in implementing FAIR data principles include fragmented data systems and formats, a lack of standardized metadata or ontologies, the high cost and time investment required to transform legacy data, cultural resistance to implementing FAIR and infrastructure not built for multi-modal data. This blog post will explore the history, purpose and importance of FAIR data principles as well as actionable ways to make data more FAIR.
What are the FAIR data principles?
FAIR data principles are four guiding rules intended to improve the Findability, Accessibility, Interoperability and Reuse (FAIR) of research data for better scientific data management and stewardship. These principles focus on preparing data for computational systems to find, access, interoperate and reuse data with minimal human intervention because of the vast complexity of today’s research data.
As life sciences and biotech research in Big Pharma embraces the potential of multi-modal data like omics, single cell, imaging and EHR, FAIR data principles have become essential to designing data infrastructure that can manage this complex data and accelerate knowledge discovery.
Who developed the FAIR data principles?
The FAIR data principles were created by Mark D. Wilkinson and many other scientists, academics and industry representatives in a 2016 article called “FAIR Guiding Principles for scientific data management and stewardship” for Scientific Data. This FAIR initiative paper lays out how they designed the FAIR Data Principles to guide researchers who wanted “to enhance the reusability of their data holdings” and improve the capacity of computational systems to automatically find and use data.
What does each FAIR principle stand for?
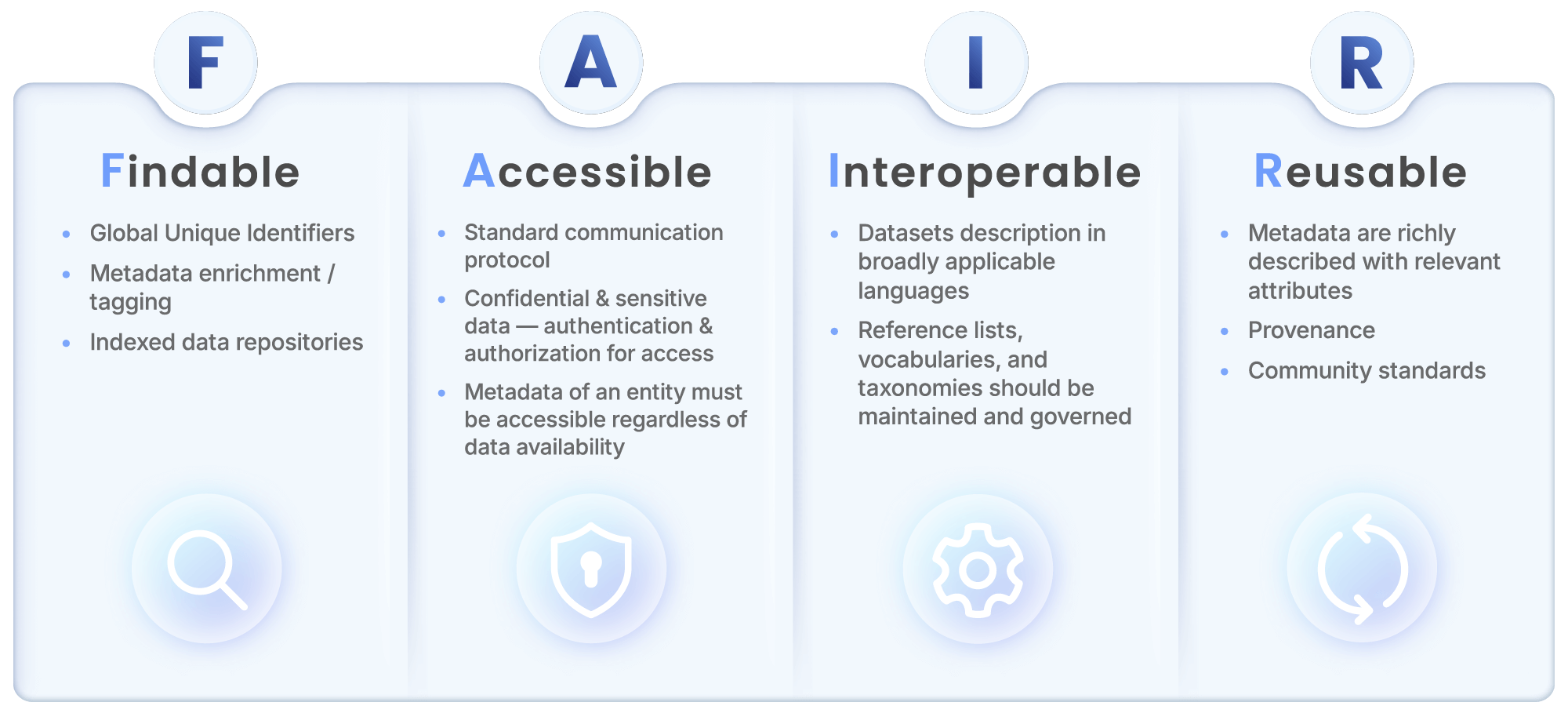
The FAIR principles are an acronym for Findable, Accessible, Interoperable and Reusable. Here’s what each principle means and its significance for life sciences research:
Findable: Data should be easy for researchers and computer systems to discover. This means assigning globally unique and persistent identifiers (such as DOIs or UUIDs) to all datasets and ensuring datasets are indexed with rich, machine-actionable metadata. The Findable principle is important because it lays the groundwork for efficient knowledge reuse by making research data, assays, and results easy to locate across departments, collaborators, and platforms.
Accessible: Data must be retrievable by users through standardized communication protocols, even if the data is restricted to select users and behind secure authentication and authorization layers. In this case, restricted data should have clear permissions so users know the path to access. The Accessible principle is key to implementing infrastructure that supports controlled data access at scale, ensuring the right people can access the right data without compromising security or compliance.
Interoperable: Data needs to be machine-readable and compatible with other systems and formats outside of the initial experimental environment. In practice, this means data must be described using standardized vocabularies and ontologies, and stored in machine-readable formats that can be seamlessly combined. The Interoperable principle is vital because multi-modal research environments rely on integrating diverse datasets like genomic sequences, imaging data and clinical trials so researchers and machine-learning technology across the world can read them.
Reusable: Data must be able to be easily replicated and studied in new contexts. This requires clarity on licensing and usage rights, robust documentation of data provenance, quality and context as well as annotating the datasets with rich and well-described metadata. Because the primary goal of FAIR is to optimize the reuse of data, the Reusable principle is vital to maximizing how useful datasets are to global researchers looking for breakthroughs.
What is the difference between FAIR data and open data?
FAIR data is focused on making data findable, accessible, interoperable and reusable, not necessarily publicly available. FAIR data principles aim to ensure data is well-structured, richly described (including indexing metadata) and machine-actionable so as to maximize data utility in complex multi-modal research environments.
Here’s an example of FAIR data in a biotech research context: A company’s internal preclinical assay results are usually governed by strict confidentiality and IP protection, which means this data is not open. However, these datasets can be FAIR if they have persistent identifiers (e.g., UUIDs), use controlled vocabularies, include rich metadata and are accessible to authorized users in well-documented APIs.
Open data is data made freely available for anyone to access, use, modify and share without restrictions. Open data is designed to serve the public good with its free accessibility, but this means it is not always compatible with patient privacy rules, IP protection and other restrictions governing data access.
Here’s an example of open data in life sciences research: The NCBI’s GenBank is the NIH’s annotated collection of all publicly available DNA sequences and follows open data principles. This means anyone can access and download the data without restrictions, but this open science data would not be FAIR unless it was properly curated with metadata and interoperable data formats.
In essence, the main difference between FAIR and open data is who is using the data: FAIR data is focused on making data easily usable by computational systems, and open data is focused on making data freely available for anyone to access it.
Why are the FAIR data principles important?
FAIR data principles are vital to life sciences and biotech research because they prepare data for machine-learning applications and other computational systems that can handle the scale of multi-modal data. Here are five key ways FAIR data principles drive value across the research lifecycle:
Enables faster time-to-insight
Locating, understanding and formatting data can cost a lot of time that researchers would rather spend in useful analysis. FAIR data speeds this time to insight by ensuring datasets are easily discoverable, well-annotated and machine-actionable.
When scientists and their data management tools can access the information they need on well-structured and annotated datasets, experiments run faster and complete sooner. This accelerates timelines for drug discovery, biomarker identification and other research outputs. A paper titled “Scientific Dataset Discovery via Topic-level Recommendation” describes how improving dataset discoverability helped researchers identify pertinent datasets more efficiently and accelerate the completion of experiments.
Improves data ROI and reduces infrastructure waste
Life sciences organizations invest a great deal in generating and storing research data, but this data is often underused because of poor organization, missing metadata and inaccessible formats. FAIR data maximizes the value of existing data assets by ensuring each dataset remains discoverable and usable throughout its lifecycle. This prevents duplication, reduces the need for repetitive training and maximizes the return on investment in both data generation and infrastructure.
Supports AI and multi-modal analytics
Multi-modal data plays a key role in drug development, but this frontier data is highly complex and difficult to understand without data management technology like AI and machine-learning applications. FAIR data provides the foundation needed to harmonize diverse data types into machine-readable formats with rich metadata that allow for algorithmic data processing, which is essential for scaling AI and ML projects in life sciences.
By making data ready for advanced analytics, machine learning and predictive modeling, FAIR facilitates research across multi-omics, imaging and EHRs. Scientists at the United Kingdom’s Oxford Drug Discovery Institute used FAIR data in databases powered by AI to speed Alzheimer’s drug discovery by reducing gene evaluation time from a few weeks to a few days.
Ensures reproducibility and traceability
Scientific integrity depends on the ability to replicate results and trace data back to its source. FAIR data supports these goals by embedding metadata, provenance and context to help research teams track how data was collected, processed and interpreted.
By ensuring reproducibility and traceability, FAIR data maintains scientific rigor and simplifies publication compliance and regulatory review. Researchers in the BeginNGS coalition accessed reproducible and traceable genomic data from the UK Biobank and Mexico City Prospective Study using query federation, helped to discover false positive DNA differences and reduce their occurrence to less than 1 in 50 subjects tested.
Enables better team collaboration across silos
How do the FAIR principles differ from CARE principles?
FAIR principles emphasize the quality, structure and utility of data by making it findable, accessible, interoperable and reusable, and the CARE principles focus on responsible data use involving Indigenous peoples and other historically marginalized populations. Developed by the Global Indigenous Data Alliance, the CARE principles stand for:
Collective benefit – Data ecosystems must be used in ways that benefit and support the well-being of Indigenous communities.
Authority to control – Indigenous peoples must have the right and power to control data related to their people, lands and cultures.
Responsibility – Anyone working with Indigenous peoples’ data must act transparently and be accountable for how this data will be used to support Indigenous peoples’ self-determination.
Ethics – Data practices must respect the cultural values and the rights of Indigenous peoples and communities across all stages of the data life cycle.
While the FAIR Principles and the CARE Principles have different priorities and requirements, the two are not mutually exclusive and are both useful for biotech researchers. Here is a quick breakdown of the primary differences between FAIR and CARE:
FAIR Principles | CARE Principles |
Focuses on data quality Enables greater machine-actionability Enhances data’s technical usability Requires data to be findable, accessible, interoperable and reusable | Focuses on data ethics Enables better cultural understanding Enhances data’s social equity and respect for Indigenous people Requires data to be for collective benefit, controllable by Indigenous people, used responsibly and ethically handled to respect Indigenous people |
What are the common challenges in implementing FAIR data principles?
FAIR data principles are considered best practices for data management in life sciences, but implementing them in real-world departments is not always easy. The complex and highly regulated nature of biotech and pharmaceutical research involves large heterogeneous datasets and many data management systems. This can make the journey toward implementing FAIR principles full of technical and organizational challenges. Here are some of the most common challenges in implementing FAIR data principles:
Fragmented data systems and formats
Example: A principal investigator is working on translational research at a biotech company. But when she tries to integrate imaging and genomics data, she has to combine a variety of clouds and storage systems from different teams and learns that each team stored their datasets in different formats. Before she can analyze all this data at scale, she has to painstakingly put all the data into interoperable formats in her data repository.
Lack of standardized metadata or ontologies
Example: A bioinformatician at a life sciences organization wants to align multi-omics datasets to inform his team’s target discovery. However, because different labs and vendors use different vocabularies for gene naming and disease codes), there are numerous semantic mismatches and ontology gaps for his team to correct. With phenotype descriptors varying between all these cohorts, their research faces significant delays.
High cost and time investment in transforming legacy data
Making legacy data FAIR often involves retrofitting decades' worth of data stored in non-standardized formats through manual metadata annotation, reformatting and conversion into new schemas. Because older data systems may lack APIs or integration capabilities, this transformation requires increasing engineering overhead for data wrangling, curation and infrastructure redesign. This is often time-consuming and challenging due to research budget constraints and limited headcount.
Example: An IT leader at a big pharma firm needs to reprocess a decade’s worth of clinical trial data from different sources. However, she learns most of this data lacks rich metadata and was stored in Excel files, scanned PDFs and other formats. Without the budget to add data engineers or more flexible data technology to her team, this data cleanup project’s estimated time-to-completion goes from three months to nine months.
Cultural resistance or lack of FAIR-awareness in teams
Example: A head of R&D IT at a life sciences company is leading a project to standardize data workflows across her organization. However, because different labs are protective of their own data formats and pipelines, she realizes each team has a different approach for file names, storage management and processing methods. As she struggles to gain FAIR buy-in throughout her firm, many researchers push back by calling her project unnecessary admin work that is slowing their discovery.
Infrastructure built for tabular, not multi-modal, data
Most data management platforms were built for data structured in relational tables and are ill-suited for frontier data like genomics, imaging, spatial omics or unstructured text. Tabular databases struggle to handle the high-dimensional or large-volume files required in research areas like single-cell or population genomics. This leads to slower analysis and increased computing costs. Implementing FAIR without flexible data infrastructure that can scale with multi-modal data is an uphill battle.
Example: A Head of Data Science at a biotech company needs to analyze imaging and transcriptomics data in a centralized location. However his existing data management platforms designed for tabular databases struggle to scale with these high-resolution datasets. His team’s current solution also can’t store both data types efficiently, which leads to time-consuming workarounds outside the central data management platform that cause additional data fragmentation.
How to make data more FAIR?
Making data Findable, Accessible, Interoperable, and Reusable is key to unlocking its long-term value for discovery, translational research and AI-driven innovation. Here are practical steps research teams in biotech and life sciences can take to pursue the FAIR guiding principles and how TileDB serves as a FAIR data principles platform in each area:
Findable
To make your data more findable, implement consistent identification with well-structured metadata and build centralized catalogs that facilitate search.
Assign persistent identifiers such as DOIs (Digital Object Identifiers) for published datasets or UUIDs (Universally Unique Identifiers) for internal records, ensuring long-term traceability for all data.
Create centralized and searchable data catalogs that make it easy for users across departments to locate datasets based on key attributes, context or modality.
Use standardized metadata schemas such as ISA-Tab, Dublin Core or MIAME for microarrays to describe datasets in a machine-readable and domain-relevant way to prepare data for AI and ML applications in research.
TileDB’s platform makes data more findable by centralizing data from diverse sources and formats and attaching rich metadata, making it easy for stakeholders across your organization to search for the datasets they need. Learn how we helped centralize a unified data catalog for Cellarity.
Accessible
To improve the accessibility of your data, you need to define access protocols clearly, store data in environments with APIs and ensure even restricted data still has metadata describing how to request access.
Define access protocols and permissions clearly by transparently communicating which users and systems can access what data and under what conditions.
Store data in environments with APIs or control access via governed interfaces (such as S3 buckets, FAIR Data Points or cloud-based research platforms) to allow secure access at scale.
Ensure even restricted datasets include rich metadata that describes the data in questions and how a user can request or gain access through appropriate channels.
TileDB’s technology makes data more accessible through asset views that facilitate access to massive datasets across teams and projects without moving or duplicating the physical data on cloud storage. Our teamspaces also control access to data by tracking and auditing access history from dataset queries and writes, including statuses, logs and resource consumption. Here’s how TileDB built a robust consent-tracking system into a new data mesh for Quest Diagnostics.
Interoperable
The key to increasing the interoperability of your data is to always ask “What will make it easier for a computational system or AI platform to understand and read this data?” Here are three interoperability best practices:
Use standardized data formats appropriate to your domain, such as VCF for genomic variants, HDF5 for high-dimensional bioimaging or JSON for structured clinical records.
Adopt shared ontologies and controlled vocabularies like those from the OBO Foundry, CDISC or SNOMED CT to ensure semantic consistency across projects.
Optimize for machine-actionability by encoding relationships, metadata and variables in structured, parsable formats rather than in free text or PDFs.
To simplify these interoperable-focused steps, TileDB’s multi-dimensional arrays natively capture the structure of all ingested data regardless of its modality, format or file type. This helps you rapidly catalog all kinds of data from local and cloud environments in bulk, making your data ready for AI and ML applications. Discover how TileDB helped Phenomic AI build a serverless cloud architecture that enables their experts to focus on scientific analysis and ML, not data engineering and pipelines.
Reusable
To make your data as reusable as possible, focus on clarifying your documentation and metadata, implementing ethical licensing and validating analyses to ensure data quality.
Apply clear licensing terms (e.g., CC-BY and CC0) and citation guidelines to help your data be used with confidence and appropriate attribution.
Include rich metadata that captures the who, what, when and how of the dataset, including the instruments used, conditions, access restrictions, transformations and assumptions.
Validate and document data quality with logs, QA metrics or provenance records so users can trust and verify findings using your documentation.
TileDB helps you make your data FAIR at source, helping your analytics also be FAIR to drive reproducibility. By supporting detailed metadata, provenance tracking and industry compliance, TileDB ensures your data is well-documented, licensed and ready for reuse in new contexts or by new users. Learn how TileDB helped Rady Children’s Hospital reduce the diagnostic burden on clinicians across the BeginNGS coalition by incorporating a wide range of annotation data.
Conclusion: TileDB for your FAIR data solution?
FAIR data principles envision a future where all research data is findable, accessible, interoperable and reusable. These data qualities are vital to life sciences research because they empower collaboration across teams and departments, improve time-to-insight and data infrastructure ROI and make it easier for advanced AI and ML applications to analyze complex multi-modal data. As your organization looks for ways to overcome the challenges of implementing FAIR data principles and go FAIR, the right database technology partner can make a huge difference.
TileDB has partnered with global life sciences firms to provide the only database designed for discovery. Our multi-dimensional arrays greatly simplify your FAIR data journey by organizing, structuring, easing collaboration on and analyzing frontier data no matter its modality. To explore how we can equip your firm with a FAIR data principles platform, contact us today.
Meet the authors

